Cornell researchers have answered a fundamental question about how two strands of DNA, known as a double helix, separate to start a process called replication, in which genes copy themselves.
The research, published in the current issue of the journal Cell, examined the role of an enzyme called a helicase, which plays a major role in separating DNA strands so that replication of a single strand can occur.
Scientists have known that helicases bind to the area of a double helix where the two strands fork away from each other, like the free ends of two pieces of thread wound around each other. The forked area opens and closes very rapidly. But scientists have debated whether helicases actively separate the two strands at the fork or if they passively wait for the fork to widen on its own.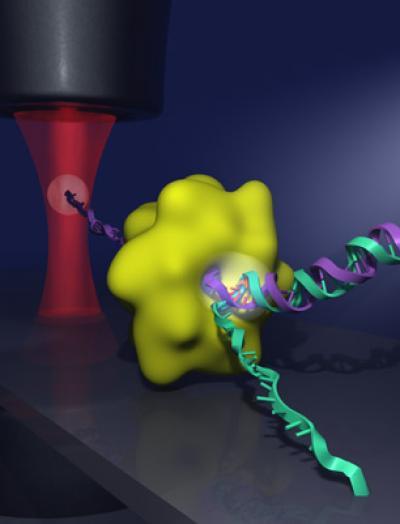 This image shows a DNA double helix (green and purple strands) being separated by a helicase enzyme (green globule) at the junction where the two strands fork. To show that helicases actively separate the two strands of DNA, the researchers attached one end of a DNA strand to a microscope cover slip and attached the end of the other DNA strand to a micron-sized plastic bead. The bead was then trapped in a tightly focused laser beam (red), which allowed the researchers to measure the motion of the helicase as it unwound the DNA. Credit: Graphic created by Chris Pelkie and Daniel Ripoll/Cornell Theory Center
This image shows a DNA double helix (green and purple strands) being separated by a helicase enzyme (green globule) at the junction where the two strands fork. To show that helicases actively separate the two strands of DNA, the researchers attached one end of a DNA strand to a microscope cover slip and attached the end of the other DNA strand to a micron-sized plastic bead. The bead was then trapped in a tightly focused laser beam (red), which allowed the researchers to measure the motion of the helicase as it unwound the DNA. Credit: Graphic created by Chris Pelkie and Daniel Ripoll/Cornell Theory Center
The research found that the helicase appears to actively exert a force onto the fork and separate the two strands.
"A simple passive unwinding mechanism does not explain our data," said Michelle Wang, associate professor of physics and the paper's senior author.
"Defects in helicases are associated with many human diseases, ranging from predisposition to cancer to premature aging," said co-author Smita Patel, a biochemistry professor at the Robert Wood Johnson Medical School in Piscataway, N.J. "Helicases are involved in practically all DNA and RNA metabolic processes."
The researchers made their discovery by anchoring one end of one of the strands in a double helix to the surface of a microscope cover slip. The end of the other strand was attached to a micron-sized plastic bead. They then focused a laser beam on the tiny bead and trapped the bead in place within the beam of light. This setup allowed the researchers to measure the position and force on the bead, creating a very precise sensor of the helicase motion. As the helicase moved toward the fork and the double helix unwound, the tension on the two strands lessened. Using statistical mechanics models, the researchers could then compare actual measurements of movement with predictions based on both active and passive scenarios.
"The unwinding has to have some active component to it, and based on our data, we can tell you exactly how active it is," said Wang. "Basically, it is an active unwinding motor."
While helicases unwind very rapidly in cells, in test tube experiments the unwinding is much slower. The researchers believe that helicases work with other enzymes, where "accessory proteins are helping the helicase out by destabilizing the fork junction," said Wang.
A major part of this work was done by Cornell physics graduate students Daniel Johnson, the lead author, Lu Bai and Benjamin Smith.
Source: Cornell