Scientists have for the first time reengineered a building block of a geometric nanocompartment that occurs naturally in bacteria. They introduced a metal binding site to its shell that will allow electrons to be transferred to and from the compartment. This provides an entirely new functionality, greatly expanding the potential of nanocompartments to serve as custom-made chemical factories.
Scientists hope to tailor this new use to produce high-value chemical products, such as medicines, on demand.
The sturdy nanocompartments, which are polyhedral shells composed of triangle-shaped sides and resemble 20-sided dice, are formed by hundreds of copies of just three different types of proteins. Their natural counterparts, known as bacterial microcompartments or BMCs, encase a wide variety of enzymes that carry out highly specialized chemistry in bacteria.
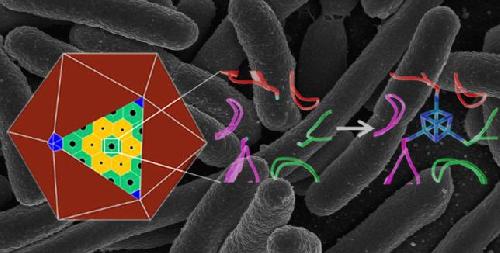 This image shows a natural atomic-scale protein structure (middle) in a polyhedral bacterial microcompartment (left), and an engineered structure (right) that binds an iron-sulfur cluster (in blue), giving it a new function. The engineered protein was produced in E. coli bacteria--the background image shows a scanning electron micrograph image of E. coli. Credit: Berkeley Lab, National Institutes of Health
This image shows a natural atomic-scale protein structure (middle) in a polyhedral bacterial microcompartment (left), and an engineered structure (right) that binds an iron-sulfur cluster (in blue), giving it a new function. The engineered protein was produced in E. coli bacteria--the background image shows a scanning electron micrograph image of E. coli. Credit: Berkeley Lab, National Institutes of Health
Researchers at the Department of Energy's Lawrence Berkeley National Laboratory (Berkeley Lab) devised synthetic shell structures derived from those found in a rod-shaped, ocean-dwelling bacterium, Haliangium ochraceum, and reengineered one of the shell proteins to serve as a scaffold for an iron-sulfur cluster found in many forms of life. The cluster is known as a "cofactor" because it can serve as a helper molecule in biochemical reactions.
BMC-based shells are tiny, durable and naturally self-assemble and self-repair, which makes them better-suited for a range of applications than completely synthetic nanostructures.
"This is the first time anyone has introduced functionality into a shell. We thought the most important functionality to introduce was the ability to transfer electrons into or out of the shell," said Cheryl Kerfeld, a structural biologist at Berkeley Lab and corresponding author in this study. Kerfeld's research group focuses on BMCs. Kerfeld holds joint appointments with Berkeley Lab's Molecular Biophysics and Integrated Bioimaging (MBIB) Division, UC Berkeley and the MSU-DOE Plant Research Laboratory at Michigan State University (MSU).
"That greatly enhances the versatility of the types of chemistries you can encapsulate in the shell and the spectrum of products to be produced," she said. "Typically, the shells are thought of as simply passive barriers."
Researchers used X-rays at Berkeley Lab's Advanced Light Source (ALS) to show, in 3-D and at the atomic scale, how the introduced iron-sulfur cluster binds to the engineered protein.
The study is now online in the Journal of the American Chemical Society.
Enzymes inside natural BMCs can convert carbon dioxide into organic compounds that can be used by the bacteria, isolate toxic or volatile compounds from the surrounding cell, and carry out other chemical reactions that provide energy for the cell.
In this study, researchers introduced the iron-sulfur cluster into the tiny pores in the shell building block. This engineered protein serves as an electron relay across the shell, which is key to controlling the chemical reactivity of substances inside the shell.
Clement Aussignargues, the lead author of the study and postdoctoral researcher in the MSU-DOE Plant Research Laboratory in Michigan, said, "The beauty of our system is that we now have all the tools, notably the crystallographic structure of the engineered protein, to modify the redox potential of the system--its ability to take in electrons (reduction) or give off electrons (oxidation).
"If we can control this, we can expand the range of chemical reactions we can encapsulate in the shell. The limit of these applications will be what we put inside the shells, not the shells themselves."
He added, "Creating a new microcompartment from scratch would be very, very complicated. That's why we're taking what nature put before us and trying to add to what nature can do."
To design the metal binding site, Kerfeld's group first had to solve the structures of the building blocks of the nanocompartment to use as the template for design. These building blocks self-assemble into synthetic shells, which measure just 40 nanometers, or billionths of a meter, in diameter. The natural form of the shells can be up to 12 times larger.
The iron-sulfur cofactor of the engineered protein, which was produced in E. coli bacteria, was very stable even when put through several redox cycles--a characteristic essential for future applications, Aussignargues said. "The engineered protein was also more stable than its natural counterpart, which was a big surprise," he said. "You can treat it with things that normally make proteins fall apart and unwind."
A major challenge in the study was to prepare the engineered protein in an oxygen-free environment to form tiny crystals that best preserve their structure and their cofactor for X-ray imaging, Kerfeld said. The crystals were prepared in an air-sealed glovebox at MSU, frozen, and then shipped out for X-ray studies at Berkeley Lab's ALS and SLAC National Accelerator Laboratory's Stanford Synchrotron Radiation Lightsource (SSRL).
In follow-up work, the research team is exploring how to incorporate different metal centers into BMC shells to access a different range of chemical reactivity, she said.
"I'm working on incorporating a completely different metal center, which has a very positive reduction potential compared to the iron-sulfur cluster," said Jeff Plegaria, a postdoctoral researcher at the MSU-DOE Plant Research Laboratory who contributed to the latest study. "But it is the same sort of idea: To drive electrons in or out of the compartment."
He added, "The next step is to encapsulate proteins that can accept electrons into the shells, and to use that as a probe to watch the electron transfer from the outside of the compartment to the inside." That will bring researchers closer to creating specific types of pharmaceuticals or other chemicals.
source: DOE/Lawrence Berkeley National Laboratory