HOUSTON - (Feb. 8, 2016) - Rice University bioengineers have found new techniques for precision genome editing that are more accurate and have fewer off-target errors.
The new strategies are shared in three papers in an upcoming special issue of the Nature journal Molecular Therapy on improving the revolutionary genome-editing technique called CRISPR-Cas9.
Bioengineering Professor Gang Bao and his colleagues present ideas for maximizing on-target gene editing with biological catalysts capable of cutting DNA called "engineered nucleases." Several such systems have been studied for years, but for the past three, the promise of cut-and-paste editing via CRISPR-Cas9 has captured the attention of scientists worldwide.
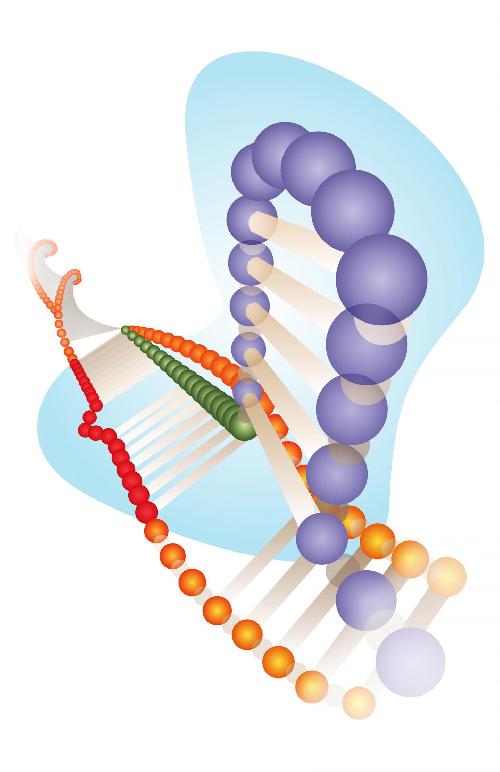 A Cas9 protein (light blue) with guide RNA (purple) and DNA (red) shows a DNA bulge, marking a sequence that would be considered off-target for CRISPR-Cas9 editing. The Rice University lab of bioengineer Gang Bao has developed Web-based tools to search for such off-targets. Credit: Bao Lab/Rice University
A Cas9 protein (light blue) with guide RNA (purple) and DNA (red) shows a DNA bulge, marking a sequence that would be considered off-target for CRISPR-Cas9 editing. The Rice University lab of bioengineer Gang Bao has developed Web-based tools to search for such off-targets. Credit: Bao Lab/Rice University
CRISPR-Cas9, a naturally occurring defense system in bacteria, allows researchers to design a short sequence of RNA called "guide RNA" that targets a specific section of genetic code (DNA) in a cell. An associated Cas9 protein then cuts the section, disrupts it or replaces it with the desired code.
That's how bacteria use CRISPR-Cas9 to immunize themselves from disease. Exposure to an invader causes the bacteria to adapt by adding the invader's genetic signature to a CRISPR database. The bacteria then recognize future enemies and destroy them with an appropriate Cas9 protein.
About three years ago researchers discovered that bacterial CRISPR-Cas9 could be modified to edit DNA in human cells by, for instance, replacing mutant sequences with normal, or "wild-type," sequences in much the same way a bacterium banks an invader's DNA signature. The technique is seen as having great potential for disease modeling and treatment, synthetic biology and molecular pathway dissection.
But CRISPR-Cas9 is still vulnerable to snipping the wrong sequences -- called "off-targets" -- in addition to the right ones. In therapeutic applications, Bao said, off-target cutting by CRISPR-Cas9 could cause many detrimental effects, including cancer.
Bao, who moved to Rice's BioScience Research Collaborative (BRC) in 2015 with a grant from the Cancer Prevention and Research Institute of Texas, is studying ways to refine CRISPR-Cas9, which he described as "nanoscissors for editing genes."
One of his goals is to treat the hereditary disease sickle cell anemia, which he hopes CRISPR-Cas9 will eventually cure. But first the therapy must become much better at avoiding off-targets that can cause unwanted side effects.
In two of the papers, the researchers study different orthologs: Cas9 proteins from species with the same ancestors as the Streptococcus pyogenes (Spy) bacterium commonly used in CRISPR/Cas9.
"Our approach in these papers is to explore the possibility of using different Cas9 orthologs," Bao said. "There are many possibilities."
In the first paper, Bao and his group used experiments on mammalian cells to characterize a CRISPR-Cas9 system from the Neisseria meningitides (Nme) bacterium. It differs from Spy in a way that bioengineers can use to reduce the risk of off-target edits, he said.
That difference lies primarily in a sequence of code that is not part of the target, but close by. Known as a protospacer-adjacent motif (PAM), it's a marker for target DNA sequences and necessary for Cas9 protein binding. In SpyCas9 editing, the PAM sequence is generally three nucleotides long. For Nme, the required PAM sequence is significantly longer -- eight nucleotides. While Nme may find fewer targets, those targets are more likely to be the correct ones, according to the researchers. That, they argue, may make it a safer alternative for gene editing.
The second paper, a collaboration with colleagues at the University of Freiburg, Germany, addresses highly specific human-gene editing using yet another bacteria's immune system. For this study, Cas9 proteins from Spy were replaced with Streptococcus thermophiles (Sth) proteins that also recognize longer PAMs. Tests carried out in human cells found Sth proteins with more stringent PAM requirements were significantly better than SpyCas9 proteins at avoiding off-targets.
Bao and company also looked at the effect of bulges in DNA and RNA that can influence targeting. Bulges appear when a sequence is one nucleotide longer or one nucleotide shorter than the expected DNA sequence targeted by guide RNA.
"We found that even with DNA or RNA bulges, the Cas9 protein can still cut," he said. "That's a unique contribution. Nobody saw that would be the case, but we demonstrated it. Consequently, we've developed a Web-based tool to search for three cases of potential off-target sites that contain base mismatches, RNA bulges and DNA bulges."
Bao noted the Nme and Sth Cas9 proteins, unlike Spy, are small enough to be packaged within an adeno-associated virus for delivery to and treatment of specific cells in an animal. "That's another advantage, and why we want to go on to explore these two systems," he said.
The third paper is a review of current CRISPR-Cas9 techniques that focuses on genome-editing tools available for target selection, experimental methods and validation. Bao and his team also lay out a list of challenges yet to be solved to eliminate off-target effects.
He said there is a path forward, represented in part by his investigation of two new bacterial systems as well as the fact that CRISPR-Cas9 is a much easier technique to implement in the lab than other genome-editing systems such as TALEN and zinc finger nuclease.
Bao said that unlike those older genome-editing techniques, CRISPR-Cas9 is straightforward enough for students to learn and use in a short time.
Bao hopes to establish his lab as a focal point for genome editing in the Texas Medical Center. To that end, he brought the TMC genome-editing community together for a well-attended workshop at the BRC last December.
"We had a lot of good discussions," he said. "One thing I would like to stimulate is the formation of a consortium among the many labs in TMC using CRISPR. They have needs to design CRISPR systems for different applications, but there are a lot of common issues. If we work together, it will be easier to address them."
source: Rice University