Like the phenomena of flocking birds and shoaling fish, the dance of molecules across a cell's surface has long fascinated theorists, physicists and biologists alike. Unlike bird and fish behaviour, however, cell surface dynamics cannot be observed and studied easily. However, it is important to understand these processes as they are crucial for cells to gain information about their environment and respond. So how does one understand the rules that govern movement of molecules across this arena? By reconstructing the cell surface from scratch, perhaps? Now, scientists from the National Centre for Biological Sciences (NCBS) in Bangalore have managed to do exactly that - construct a simplified cell surface from its constituent parts, namely, a mixture of fats and proteins. This reconstruction creates a crucial new tool that researchers can use to test theories on cell surface dynamics.
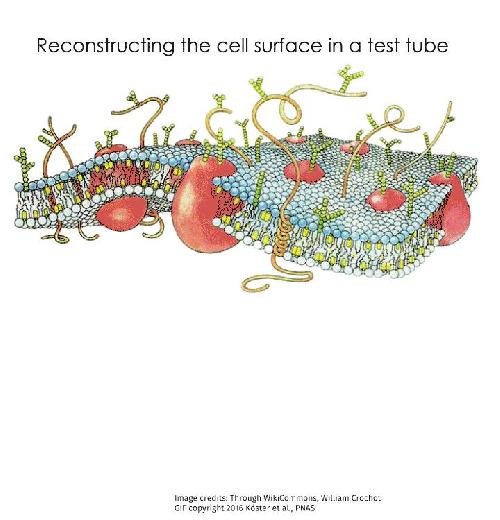
Molecular movements on the cell surface are known to be non-random, incredibly complex, and do not seem to follow simple thermodynamic rules. Until recently, there were few experimental tools available to study such phenomena to really understand how the cell surface functioned. This has changed with the new experimental system that has been developed by a close collaboration between experimental biologists from Prof. Satyajit Mayor's group at NCBS, scientists from the University of California San Francisco (UCSF) and theoretical physicists from Prof. Madan Rao's group at NCBS and RRI (Raman Research Institute). The experimental system is a minimal model of the cell surface constructed from its basic components - purified fats and proteins known to be part of the cell surface. This tool could be the key to understanding how the surface of a living cell works.
"This is just a beginning but an important one," says Prof Satyajit Mayor. "Important because it allows one to test ideas that have come from theory built around providing an explanation for active organization at the surface of a living cell. It's an exciting beginning since the feasibility of this simple minimal system opens up huge possibilities to explore the world of a living cell in a test tube system where every element is under our control. This work is inspired by the adage 'what we understand we should be able to build' and this is in trying to understand the principles behind how a living material, the cell surface, works," he adds.
 Three channel time lapse of myosin-induced actin contraction into polar asters. (Scale bar: 10 µm). Credit: Image credits: Through WikiCommons, William CrochotGIF copyright 2016 Köster et al., PNAS
Three channel time lapse of myosin-induced actin contraction into polar asters. (Scale bar: 10 µm). Credit: Image credits: Through WikiCommons, William CrochotGIF copyright 2016 Köster et al., PNAS
The 'active composite model' of the cell surface is one of the latest theories that attempts to explain the behaviour of cell surface molecules. This model visualises the cell surface as not just the cell membrane, but as an amalgamation of two elements - the cell membrane, made of fats and an interwoven mesh of the protein 'actin' that forms a thin layer just below the cell membrane. Another protein, 'myosin' that interacts with actin, behaves like a molecular motor and creates movement in the actin meshwork when supplied with energy. Many of the cell surface proteins whose movements have baffled scientists are often linked to the dynamic actin meshwork that lies just below the cell membrane.
As proposed by the active composite model, the researchers decided to recreate a cell surface as an assembly of a fat-based membrane and an actin meshwork. This artificial cell surface was therefore constructed using a fat bilayer, actin and a fluorescent protein specially designed to be embedded in the membrane while also being linked to actin. Using various microscopic techniques, the group was able to study the behaviour of the construct via the patterns formed by the fluorescent proteins. As predicted by the 'active composite model', the dynamics of actin-bound fluorescent proteins were found to be dependent on the dynamics of the actin meshwork. When the molecular motor, myosin, was added and chemical energy provided, the forces generated by actin-myosin interactions drove the movements of these proteins. When the chemical energy was exhausted, the actin-bound proteins aggregated to form distinct bundle or aster-like structures based on the organisation of the actin meshwork.
"The importance of active or energy consuming processes in understanding biological phenomena is becoming more and more evident. This is an emerging field in biology called 'active mechanics'. Often, the emerging organisation of biological molecules are not clear, and theoretical explanations for such observations are also far from complete. This makes it important to have proper experimental tools that go hand in hand with theory to test and improve our understanding of such systems. Our current study describes the creation of an experimental system that will serve us in this," says Darius Köster, the lead author of the study that was published in the leading journal PNAS (Proceedings of the Natural Academy of Sciences of the United States of America).
"The motivation behind this work is to analyse mechanisms influencing the dynamics and organisation of molecules on the cell surface," says Kabir Husain, another author in this study. Processes like cell growth, division, immune recognition and many others are dependent on the organisation of protein receptors and other associated molecules on the cell surface. This implies that the ability of the cell to reliably control the organisation of its surface molecules is crucial to its survival and function. With the recreation of the cell surface in a testube, scientists have gained a solid experimental footing in the race to comprehend the mechanics of cell surface organisation.
source: National Centre for Biological Sciences