Researchers at the RIKEN Omics Science Center (OSC) have developed a robotic workflow for sample preparation on the HeliScope single molecule sequencer which drastically reduces sample preparation time to from 42 days to only 8 days. The workflow uses Cap Analysis of Gene Expression (CAGE), a unique method developed at the OSC for determining transcriptional starting sites in the genome and their expression levels.
A key obstacle in DNA sequencing is the long time required for sample preparation, especially when compared with how fast next-generation sequencers are able to sequence DNA. Now, researchers at the OSC, who previously adapted the CAGE method to the HeliScope single molecule sequencer, have reduced that time using a new automated sample preparation system. Using the new system, sample preparation of 96 CAGE cDNA libraries, which would normally take a manual operator 42 days, is done in only 8 days.
The researchers further demonstrated, through a comparison of results from the analysis of transcriptional starting sites and their expression levels using CAGE on the HeliScope single molecule sequencer, that results using the new preparation system are reproducible and comparable to results using manual preparation. The new automated cDNA preparation system can also be applied to a variety of other methods and sequencers, including CAGE on the Illumina/SOLD platform, RNA-seq and full-length cDNA generation.
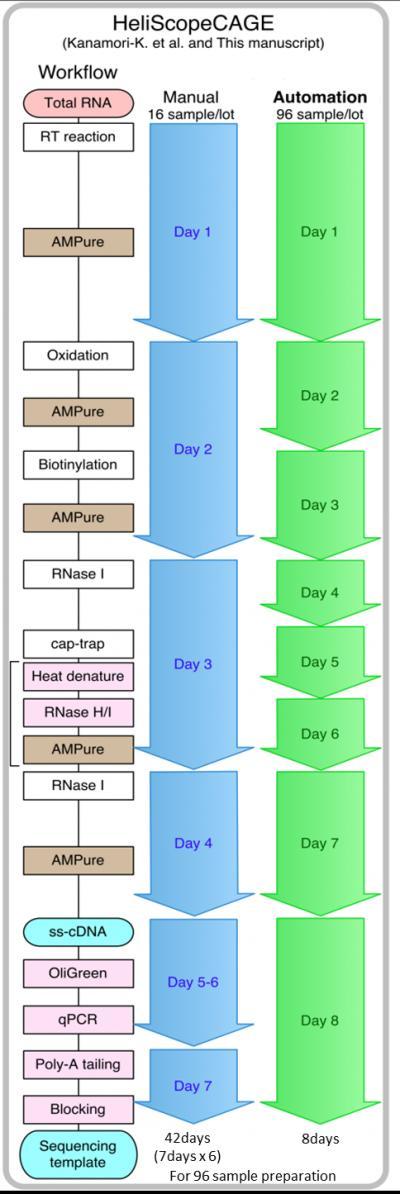
Figure1: This shows the comparison of time frames between manual and automated processes.
(Photo Credit: RIKEN)
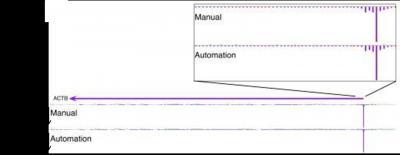
Figure 2: This shows the genomic view of a selected gene for comparison between manual and automatedmethods.CAGE mapped read counts were displayed as linear scale histogram on ACTB gene. Thetranscription initiation regions are magnified and demonstrate that the distribution oftranscription starting sites and their expression levels are consistent between manual andautomated methods.
(Photo Credit: RIKEN)
Source: RIKEN