(Boston) -- Many biological and pathological processes are not strictly controlled by the presence, absence or function of biomolecules such as proteins or nucleic acids but rather by subtle changes in their numbers at specific locations within cells. However, despite the recent revolution of optical imaging technologies that has enabled the distinction of molecular targets residing less than 200 nm apart from each other, modern super-resolution techniques still face the challenge to accurately and precisely count the number of biomolecules at cellular locations.
A new analytical tool developed by a team at the Wyss Institute for Biologically Inspired Engineering solves this problem. The team led by Peng Yin, Ph.D., a Core Faculty member at the Wyss Institute and Professor of Systems Biology at Harvard Medical School, has forged ahead with its previously developed DNA-PAINT and Exchange-PAINT super-resolution microscopy platform to now count different molecular species in biological samples with high accuracy and precision. DNA-PAINT affords higher resolution than costly super-resolution microscopes and Exchange-PAINT can survey multiple different molecules in the same biological sample. The method is reported in the March 28 issue of Nature Methods.
"We now have enhanced our DNA-powered super-resolution microscopy methods with a highly quantitative analytical tool kit. qPAINT, as we named it, can accurately count the actual numbers of specific molecules at specific locations inside the cell," said Yin. "Introducing this quantitative power has crucially extended the spectrum of imaging capabilities of this comprehensive and inexpensive technology so that it can be applied in many areas of biological and clinical research."
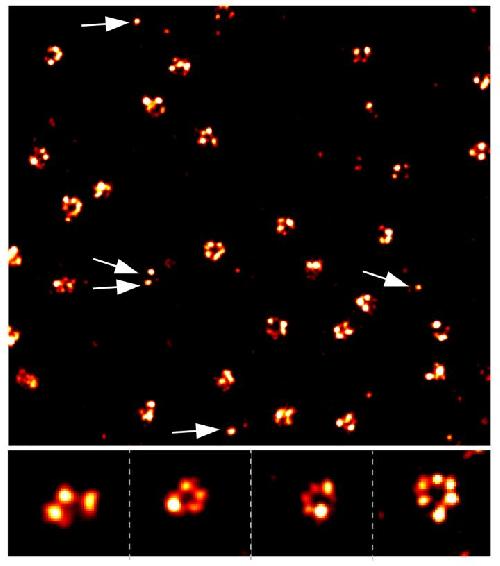 With super resolution microscopy and qPAINT analysis, researchers will be able to quantify individual molecules at specific locations in the cell. These images show varying copy numbers (shown are three, four, five, and six in the bottom images) of a protein residing in small so-called nuclear pore complexes that permit shuttling of various molecules in and out of the cell's nucleus. The arrows indicate pores with only a single protein that serve to calibrate the counting method. Credit: Wyss Institute at Harvard University.
With super resolution microscopy and qPAINT analysis, researchers will be able to quantify individual molecules at specific locations in the cell. These images show varying copy numbers (shown are three, four, five, and six in the bottom images) of a protein residing in small so-called nuclear pore complexes that permit shuttling of various molecules in and out of the cell's nucleus. The arrows indicate pores with only a single protein that serve to calibrate the counting method. Credit: Wyss Institute at Harvard University.
Key to the DNA-driven imaging technology is the transient interaction of two short strands of DNA, one called the "docking strand" that is attached to the molecular target to be visualized and the other, called the "imager strand", which carries a light-emitting dye.
"We can precisely program the time interval for which the two complementary DNA strands transiently interact with each other so that when the pair of strands goes through binding and dissociation, the dye will blink at a specific frequency. From an increase of this frequency, we can then deduce with qPAINT analysis how many targets exactly are located at a specific cellular location without spatially resolving each target," said Ralf Jungmann, Ph.D., one of the two co-first authors of the study, a former Postdoctoral Fellow in Yin's lab and now a Group Leader at the Max Planck Institute of Biochemistry at the Ludwig Maximilian University in Munich, Germany.
Applying this kind of binding analysis to DNA-PAINT and Exchange-PAINT allows the Wyss team to disregard common problems that fluorescent dyes pose for achieving truly quantitative potential in super-resolution microscopy, like their hard-to-model photophysical properties and tendency to wane under the influence of light, a phenomenon known as photobleaching.
In earlier proof-of-principle studies, the team integrated DNA-powered super resolution microscopy with highly specific and broadly available detection reagents, for example, by attaching docking strands to antibodies that specifically bind molecules in various cellular structures and complexes or to DNA probes that bind to specific messenger RNA molecules shuttling genetic information inside cells.
"With qPAINT, we counted the numbers of proteins targeted by antibodies at sites such as a cell surface, or the membrane that surrounds the cell nucleus, and even at the nerve endings that stimulate muscles to twitch. The technology can be integrated with a large array of detection reagents to eventually count diverse molecules of interest at the cellular sites where they perform their tasks," said Maier Avendaño, Ph.D., the work's other co-first author and Postdoctoral Fellow in Yin's team.
"qPAINT adds a powerful new tool to this simple super-resolution microscopy platform, which now gives researchers the extraordinary capability to quantify how changes in molecule numbers at specific locations influence cell signaling and function. And what is most amazing is that they do this without requiring a highly expensive microscope, and so it should be useable by virtually any biological or clinical laboratory," said Wyss Institute Founding Director Donald Ingber, M.D., Ph.D., who is also the Judah Folkman Professor of Vascular Biology at Harvard Medical School and the Vascular Biology Program at Boston Children's Hospital and Professor of Bioengineering at the Harvard John A. Paulson School of Engineering and Applied Sciences.
source: Wyss Institute for Biologically Inspired Engineering at Harvard