PHILADELPHIA - A team of researchers from the Perelman School of Medicine at the University of Pennsylvania have shed new light on how the structure of regulatory sequences in DNA is packaged in a cell. "This work has implications for better understanding the role that gene sequences called enhancers play within our DNA for governing gene activity," said senior author Ken Zaret, PhD, a professor of Cell and Developmental Biology and director of the Institute for Regenerative Medicine. The findings are published this week in Molecular Cell.
Enhancers influence which genes are "on" or "off" in each type of cell and also control how much each the gene is activated. This level of control ultimately dictates the quantity of an encoded protein that is made in response to particular physiological conditions.
"Recent studies have shown that errors in enhancer function are crucial in various diseases, including many types of cancers," Zaret said. "Because of this, we need to detail how enhancers work."
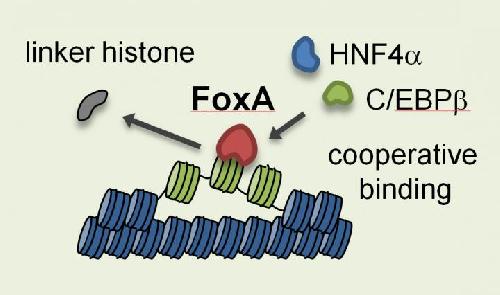 Schematic of opening of nucleosome in liver cell, with linker histone ejected, accessible nucleosomes with pioneer factors, and liver-specific transcription factors. Credit: Ken Zaret, Ph.D., Perelman School of Medicine, University of Pennsylvania
Schematic of opening of nucleosome in liver cell, with linker histone ejected, accessible nucleosomes with pioneer factors, and liver-specific transcription factors. Credit: Ken Zaret, Ph.D., Perelman School of Medicine, University of Pennsylvania
Nucleosomes, the protein complexes around which DNA in every cell winds, inherently inhibit the binding of gene regulatory proteins to chromosomes. This packaging helps keep genes in the "off" position when needed. Most researchers have worked on the assumption that nucleosomes are absent at active enhancer sites along the genome, and therefore would not play a role in enhancer function.
However, when first author Makiko Iwafuchi-Doi, PhD, a postdoctoral fellow in the Zaret lab, modified the nucleosome mapping technique, she demonstrated that nucleosomes are present, at least in part, at enhancer sites, when gene regulatory proteins called pioneer factors are attached to them. This information provides a new view of how enhancer sequences and nucleosomes interact.
To elicit the necessary physiological programming or reprogramming in a cell, gene regulatory factors must be able to engage genes that are "off" and not regularly meant for expression in cells, a finding that the Zaret lab established in a 2015 Cell paper. These silenced genes are typically embedded in tightly coiled, "closed" areas covered by nucleosomes. Transcription factors with the highest reprogramming activity have the necessary capability to interact with target sites on closed nucleosome DNA. These transcription proteins are the pioneer factors and named such because they initiate molecular changes in closed chromatin.
The team on the Molecular Cell paper went on to show that pioneer factors help expose stretches of DNA on individual nucleosomes to which they are bound. By kicking out a repressive protein called a linker histone, the pioneer factors allow other regulatory factors to gain access to the DNA. This, in turn, allows enhancers to activate genes and promote normal cell functions.
Together, these studies reveal novel insights into the ways that pioneer factors initiate and maintain enhancer sites in an active, or "on," state. The pioneer factors play important roles in controlling how different cells develop in an embryo. They also play a role in allowing genes to be responsive to hormones, including hormone-responsive human breast and prostate cancers.
"We anticipate that by understanding the basic mechanisms by which pioneer factors control enhancer function, we will be able to enhance the process by which we can change one type of cell into another," Zaret said. "This will be helpful in generating new models of human disease and, in the future, new cells for therapeutics."
source: University of Pennsylvania School of Medicine