Scientists from the U.S. Department of Energy's Lawrence Berkeley National Laboratory (Berkeley Lab) have shown for the first time that an enzyme can be tweaked to reduce lignin in plants. Their technique could help lower the cost of converting biomass into carbon-neutral fuels to power your car and other sustainably developed bio-products.
Lignin is a polymer that's important to a plant's health and structure. But lignin also permeates plant cell walls and surrounds the sugars trapped inside. This poses a major challenge in efforts to use microbes to ferment the sugar into useful chemicals and fuels. That's because lignin must be chemically broken down in a costly pretreatment step before the sugar is fermented. The less lignin there is in a plant, the cheaper this pretreatment step becomes, which is a major goal of the bioenergy industry.
Now, as reported in the journal Plant & Cell Physiology, Berkeley Lab scientists are taking on this challenge in an entirely new way.
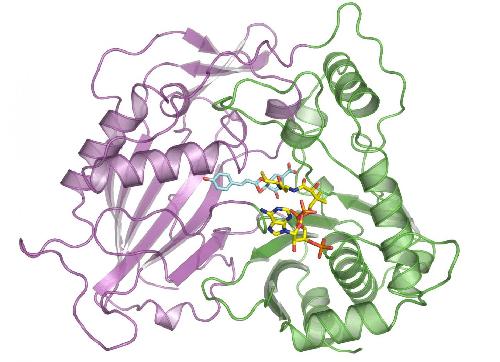 This illustration shows the molecular structure of HCT that was derived at Berkeley Lab's Advanced Light Source. The purple and green areas are two domains of the enzyme, and the multi-colored structures between the two domains are two molecules (p-coumaryl-shikimate and HS-CoA) in the binding site. New research shows this binding site is indiscriminate with the acceptor molecules it recruits, including molecules that inhibit lignin production. Credit: Berkeley Lab
This illustration shows the molecular structure of HCT that was derived at Berkeley Lab's Advanced Light Source. The purple and green areas are two domains of the enzyme, and the multi-colored structures between the two domains are two molecules (p-coumaryl-shikimate and HS-CoA) in the binding site. New research shows this binding site is indiscriminate with the acceptor molecules it recruits, including molecules that inhibit lignin production. Credit: Berkeley Lab
They focused on an enzyme called HCT that plays a key role synthesizing lignin in plants. Ordinarily, the enzyme binds with a particular molecule as part of the lignin-production process. The scientists explored whether HCT binds with several other molecules that have similar structures to the original molecule, and they found HCT is pretty indiscriminate with what it accepts.
With this in mind, the researchers introduced another molecule to the enzyme that occupies the binding site usually occupied by the lignin-producing molecule. This swap inhibits the enzyme's ability to support lignin production.
Initial tests in a model plant show this approach decreases lignin content by 30 percent while upping sugar production. What's more, the technique promises to be much more "tunable" than the current way of reducing plant lignin, in which lignin-producing genes are silenced. This decreases lignin everywhere in a plant and throughout its lifespan, resulting in a weakened plant and a lower sugar yield.
"Our goal is to tune the process so that lignin is reduced in a plant where we want it reduced, such as in tissues that produce thick cell walls, and when we want it reduced, such as later in a plant's development," says Dominique Loque, a plant biologist with the Joint BioEnergy Institute (JBEI), a DOE Bioenergy Research Center led by Berkeley Lab, which pursues breakthroughs in the production of cellulosic biofuels.
"This would result in robust bioenergy crops with more sugar and less lignin, and dramatically cheaper pretreatment costs," Loque said.
Loque conducted the research with Aymerick Eudes and several other JBEI scientists across three divisions: Feedstocks, Fuels Synthesis, and Technology. Many of the scientists are also in Berkeley Lab's Biological Systems and Engineering Division.
The scientists started their research knowing that HCT is key to lignin production in plants, and that it is potentially "promiscuous," meaning it binds with a variety of molecules. But they didn't know how promiscuous HCT is, and the extent to which HCT's promiscuity is found throughout plant life.
To find out, they isolated HCT enzymes from five plants that represent 450 million years of evolution, from ancient moss to poplar, switchgrass, pine, and a plant called Arabidopsis. They placed the enzymes in yeast and supplied the yeast with a set of new molecules suspected to bind with the enzyme instead of a lignin-producing molecule. In each case, the enzyme yielded a product that isn't an intermediate to lignin.
"This means our lignin-reduction strategy is likely applicable to most bioenergy crops," says Eudes.
Next, the scientists analyzed the enzyme's molecular structure at the Advanced Light Source, a DOE Office of Science User Facility hosted at Berkeley Lab. Protein crystallography studies of HCT from switchgrass showed one of the new molecules attaching to the enzyme at the site normally occupied by the lignin-producing molecule. In fact, the new molecule has a similar affinity to the binding site as the lignin-producing molecule.
"This is proof the enzyme accepts the new substrate, and it shows there's stiff competition between the two molecules," says Eudes.In a final test of their approach, the scientists genetically modified several Arabidopsis plants so they had 25 times more of one of the new molecules than normal. These plants had 30 percent less lignin than unmodified plants, and a higher concentration of sugars.
The scientists next want to learn how to adjust the temporal and spatial specificity of the enzyme's lignin-reduction abilities in plants. They also want to further study the Advanced Light Source-derived enzyme structures to see if HCT can be modified to be even more attractive to the new molecules.
source: DOE/Lawrence Berkeley National Laboratory