A new computational tool successfully tested in a small pilot trial harnesses clinical data to predict the optimal drug dose for an individual. The mathematical approach underlying the tool promises to take the educated guesswork out of prescribing medications for immunosuppression, cancer, heart disease, bacterial infections, and other conditions that require tightly controlled treatment regimens. Many factors including age, ethnicity, genetics, and comorbidity can influence how a patient responds to a given drug, making a "one-size-fits-all" approach to dosing inadequate. This is especially true for organ transplant patients, who are heavily treated after the operation to make sure that their body does not reject the new organ. Immunosuppressive drugs often have a narrow therapeutic range, meaning that doses above this range risk toxicity, but doses that dip below it may fail to keep transplant rejection in check. Currently, clinicians closely monitor patients and make an educated guess about adjusting the dose according to their response to the immunosuppressant drug and to the addition of other medications or therapies, like dialysis. To better guide personalized drug dosing, Ali Zarrinpar and colleagues devised an approach called parabolic personalized dosing (PPD) that harnesses an individual's clinical data, including the concentration of a drug in the bloodstream, to predict the next optimal drug dose for that patient. Unlike previous approaches based on mathematical modeling, PPD uses an algebraic equation to construct a parabolic map of a patient's response to a drug. In a pilot trial of liver transplant patients, four patients treated with an immunosuppressant drug using doses determined by PPD stayed within the target drug dosing range more frequently than did four patients managed with standard physician-guided dosing. The former group also tended to have shorter hospital stays than the latter. The researchers further showed that the approach could potentially optimize dosing for patients treated with a combination of drugs. With further development, clinicians may one day be able to plug clinical data into PPD to automatically tailor drug dosing for a wide range of patients, including those with cancer, infections, and heart disease.
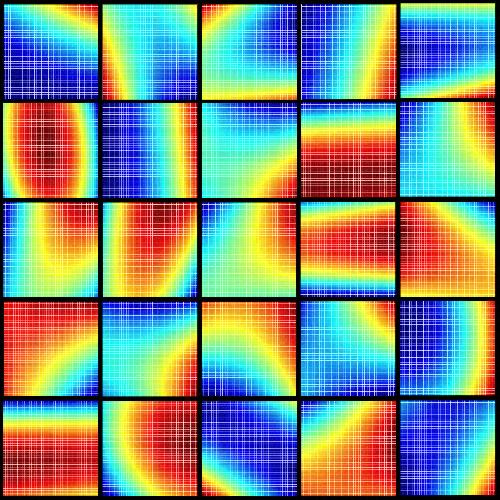 This image represents a compilation of schematic, patient-specific, parabolic response surfaces, which are the keys to personalized medicine. They represent responses to combination therapy for individual patients for liver transplant immunosuppression. The need for personalized medicine is shown by the substantial differences between individualized drug interactions. This material relates to a paper that appeared in the 6 April 2016 issue of Science Translational Medicine, published by AAAS. The paper, by A. Zarrinpar at University of California, Los Angeles in Los Angeles, Calif., and colleagues was titled, "Individualizing liver transplant immunosuppression using a phenotypic personalized medicine platform." Credit: Dr. Dong-Keun Lee, Dean Ho Group, UCLA
This image represents a compilation of schematic, patient-specific, parabolic response surfaces, which are the keys to personalized medicine. They represent responses to combination therapy for individual patients for liver transplant immunosuppression. The need for personalized medicine is shown by the substantial differences between individualized drug interactions. This material relates to a paper that appeared in the 6 April 2016 issue of Science Translational Medicine, published by AAAS. The paper, by A. Zarrinpar at University of California, Los Angeles in Los Angeles, Calif., and colleagues was titled, "Individualizing liver transplant immunosuppression using a phenotypic personalized medicine platform." Credit: Dr. Dong-Keun Lee, Dean Ho Group, UCLA
source: American Association for the Advancement of Science