The chemical bond between carbon and fluorine is one of the strongest in nature, and has been both a blessing and a curse in the complex history of fluorocarbons. Now, in a powerful demonstration of the relatively new field of "computational chemistry," researchers at the National Institute of Standards and Technology (NIST) and the Interdisciplinary Network of Emerging Science and Technology group (INEST, sponsored by Philip Morris USA) have designed—in a computer—a wholly theoretical molecule to pull the fluorine out of fluorocarbons.*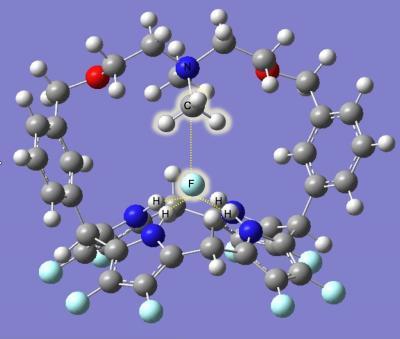 Postmortem: Computer modeling rendition of the proposed carbon-fluorine bond-breaking macrocycle after reaction with a molecule of methyl fluoride (CH3F). Highlighted in the center of the macrocycle, the CH3 fragment has attached to a nitrogen atom, separating it from the fluorine atom which has been grabbed by a group of four hydrogen atoms. The potentially toxic components of the fluorocarbon are immobilized in the macrocycle until removed by a second reaction, an important feature for possible filtering systems.
Postmortem: Computer modeling rendition of the proposed carbon-fluorine bond-breaking macrocycle after reaction with a molecule of methyl fluoride (CH3F). Highlighted in the center of the macrocycle, the CH3 fragment has attached to a nitrogen atom, separating it from the fluorine atom which has been grabbed by a group of four hydrogen atoms. The potentially toxic components of the fluorocarbon are immobilized in the macrocycle until removed by a second reaction, an important feature for possible filtering systems.
Credit: NIST
At sea level, the strong C-F bond makes fluorocarbons thermally and chemically stable. As a result, fluorocarbons have been used in many commercial applications including refrigerants, pesticides and non-stick coatings. In the upper atmosphere, however, high-energy photons and highly reactive ozone molecules can break apart fluorocarbons, with the well-known consequence of a depleted ozone layer and increased ultraviolet radiation at ground level. A determined chemist can break down fluorocarbons at ground level with certain organometallic compounds, but the reactions take a long time at very high temperatures. Other known reagents are both highly toxic and inefficient, so chemists have been searching for an economical and environmentally friendly method to dispose of fluorocarbons.
Reasoning that the problem already may have been solved by nature, the NIST/Philip Morris team looked to an enzyme called fluoroacetate dehalogenase used by a South African bacterium, Burkholderia sp. The enzyme enables the bacterium to pull the fluoride ion out of sodium fluoroacetate (disrupting a poisonous compound) at room temperature and without problematic metal ions. Enzymes are giant molecules, evolved to survive and work in the complex environment of a living organism; they can be difficult and expensive to adapt to an industrial process. Instead, the research team applied basic quantum mechanical theory of electron structures in molecules, together with the example of a known molecule that binds to and extracts chlorine ions, to calculate the make-up and geometry of the critical "active site" in the enzyme that does the work. They then designed in software a large ring-shaped molecule to hold those components in just the right orientation to break the C-F bond in methyl fluoride, a simple fluorocarbon.
Researchers at the University of Texas now are synthesizing the new molecule to test its effectiveness. If it matches theoretical predictions, it will be the first example of a simple organic molecular system able to break C-F bonds without extreme temperature and pressure conditions, and a demonstration of a novel technique for designing man-made molecules that can mimic the extraordinary selectivity and chemical activity of natural enzymes. Notes lead researcher Carlos Gonzalez, "All of these useful things are in nature, you just have to find them and make them more efficient."
* F. Hæffner, M. Marquez and C. Gonzalez. Theoretical evidence for C-F bond activation by a fluoro-calix[4]pyrrole-tert-amine macrocycle. J. Phys. Chem. A 2007, 111, 268-272.
Written from a news release by NIST.