BLOOMINGTON, Ind. -- The discovery of two genes that encode copper- and sulfur-binding repressors in the hospital terror Staphylococcus aureus means two new potential avenues for controlling the increasingly drug-resistant bacterium, scientists say in the April 15, 2011 issue of the Journal of Biological Chemistry.
"We need to come up with new targets for antibacterial agents," said Indiana University Bloomington biochemist David Giedroc, who led the project. "Staph is becoming more and more multi-drug resistant, and both of the systems we discovered are promising."
The work was a collaboration of members of Giedroc's laboratory, and that of Vanderbilt University School of Medicine infectious disease specialist Eric Skaar, and University of Georgia chemist Robert Scott.
MRSA, or multidrug-resistant Staphylococcus aureus, is the primary cause of nosocomial infections in the United States. About 350,000 infections were reported last year, about 20 percent of which resulted in fatalities, according to the Centers for Disease Control. One to two percent of the U.S. population has MRSA in their noses, a preferred colonization spot.
One of the repressors the scientists discovered, CsoR (Copper-sensitive operon Repressor), regulates the expression of copper resistance genes, and is related to a CsoR previously discovered by the Giedroc group in Mycobacterium tuberculosis, the bacterium that causes tuberculosis in humans. When the bacterium is exposed to excess copper, the repressor binds copper (I) and falls away from the bacterial genome to which it is bound, making it possible for the copper resistance genes to be turned on. This makes sense, since in the presence of a lot of copper -- a metal commonly used to kill bacteria -- a bacterium is well served by expressing genes that help the bacterium sequester and export extra copper before the metal can do any real damage.
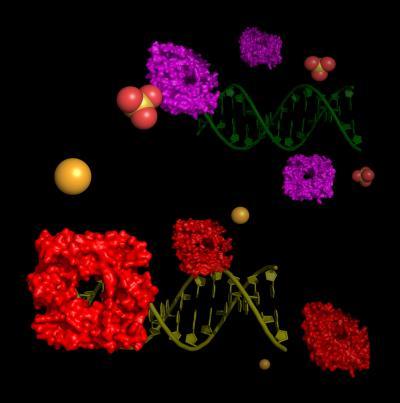
Staphylococcus aureus encodes a DNA binding copper-sensitive operon repressor (CsoR, bottom) and a CsoR-like sulfur transferase repressor (CstR, top), which are very similar to one another. Unlike CsoR, the repressor CstR does not form a stable complex with copper, Cu(I). Instead, operator binding is inhibited by attaching a second repressor to the first, possibly via a disulfide or even trisulfide bridge.
(Photo Credit: David Giedroc)
The other repressor, CstR (CsoR-like sulfurtransferase Repressor), which the scientists found can react with various forms of sulfur, appears to prevent the transcription of a series of sulfur assimilation genes based on their homology with similar genes in other bacterial species. One of the genes in this system encodes a well known enzyme, sulfurtransferase, which interconverts sulfite (SO3 2-) and thiosulfate, (S2O3 2-).
The scientists have yet to confirm the functions of the other genes controlled by CstR, but a new four-year, $1.1 million grant from the National Institutes of Health to principal investigator Giedroc will fund crucial investigations into Staph's utilization of sulfur, an important element that bacteria -- and all organisms for that matter -- use to make protein.
The two repressors -- and the gene systems they regulate -- are possible new drug targets for controlling Staph growth. A drug could hypothetically target either of the repressors, causing bacteria to become unresponsive to toxic copper levels or incapable of properly integrating sulfur into their cell physiologies, respectively.
"One thing you could do is prevent the repressors from coming off the DNA in the first place," Giedroc said "although I think that's probably a long shot. I think the repressors are one step removed from where you'd like to have the action. At this point I think the better targets are going to be the genes they are regulating."
Among those genes, Giedroc says he's hopeful one of the sulfur utilization genes controlled by CstR turns out to be an effective drug target. And he wouldn't be surprised if that was the case.
"The metabolic process by which sulfur is assimilated is a proven drug target in Mycobacterium tuberculosis," Giedroc said. "We see no reason why this can't be the case for Staphylococcus aureus. Finding out will be one of the goals of this new NIH-funded project."
Source: Indiana University