University of California, Berkeley, researchers have made a major improvement in CRISPR-Cas9 technology that achieves an unprecedented success rate of 60 percent when replacing a short stretch of DNA with another.
The improved technique is especially useful when trying to repair genetic mutations that cause hereditary diseases, such as sickle cell disease or severe combined immune deficiency. The technique allows researchers to patch an abnormal section of DNA with the normal sequence and potentially correct the defect and is already working in cell culture to improve ongoing efforts to repair defective genes.
"The exciting thing about CRISPR-Cas9 is the promise of fixing genes in place in our genome, but the efficiency for that can be very low," said Jacob Corn, scientific director of the Innovative Genomics Initiative at UC Berkeley, a group that focuses on next-generation genome editing and gene regulation for lab and clinical application. "If you think of gene editing as a word processor, we know how to cut, but we need a more efficient way to paste and glue a new piece of DNA where we make the cut."
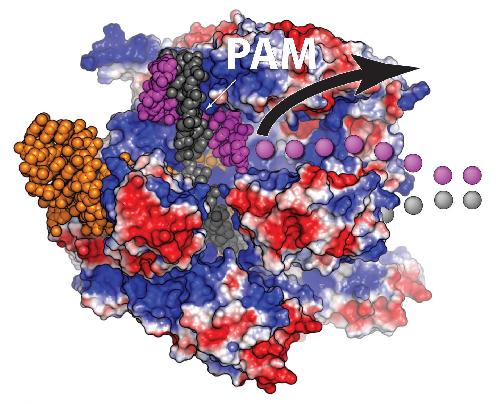 A view of the Cas9 protein (red and blue) bound to a double strand of DNA (purple and grey). After both strands are cut, one DNA strand (purple dots) is free and able to bind with a piece of DNA to be inserted at the break.This behavior can be utilized to significantly boost the efficiency of gene editing. Credit: Christopher Richardson, UC Berkeley
A view of the Cas9 protein (red and blue) bound to a double strand of DNA (purple and grey). After both strands are cut, one DNA strand (purple dots) is free and able to bind with a piece of DNA to be inserted at the break.This behavior can be utilized to significantly boost the efficiency of gene editing. Credit: Christopher Richardson, UC Berkeley
"In cases where you want to change very small regions of DNA, up to 30 base pairs, this technique would be extremely effective," said first author Christopher Richardson, an IGI postdoc.
Problems in short sections of DNA, including single base-pair mutations, are typical of many genetic diseases. Base pairs are the individual building blocks of DNA, strung end-to-end in a strand that coils around a complementary strand to make the well-known helical, double-stranded DNA molecule.
Richardson, Corn and their IGI colleagues describe the new technique in the Jan. 21 issue of the journal Nature Biotechnology.
Grabbing onto a loose strand
Richardson invented the new approach after finding that the Cas9 protein, which does the actual DNA cutting, remains attached to the chromosome for up to six hours, long after it has sliced through the double-stranded DNA. Richardson looked closely at the Cas9 protein bound to the two strands of DNA and discovered that while the protein hangs onto three of the cut ends, one of the ends remains free.
When Cas9 cuts DNA, repair systems in the cell can grab a piece of complementary DNA, called a template, to repair the cut. Researchers can add templates containing changes that alter existing sequences in the genome -- for example, correcting a disease-causing mutation.
Richardson reasoned that bringing the substitute template directly to the site of the cut would improve the patching efficiency, and constructed a piece of DNA that matches the free DNA end and carries the genetic sequence to be inserted at the other end. The technique worked extremely well, allowing successful repair of a mutation with up to 60 percent efficiency.
"Our data indicate that Cas9 breaks could be different at a molecular level from breaks generated by other targeted nucleases, such as TALENS and zinc-finger nucleases, which suggests that strategies like the ones we are using can give you more efficient repair of Cas9 breaks," Richardson said.
The researchers also showed that variants of the Cas9 protein that bind DNA but do not cut also can successfully paste a new DNA sequence at the binding site, possibly by forming a "bubble" structure on the target DNA that also acts to attract the repair template. Gene editing using Cas9 without genome cutting could be safer than typical gene editing by removing the danger of off-target cutting in the genome, Corn said.
source: University of California - Berkeley