Inorganic phosphate is an essential building block of cell membranes, DNA and proteins. It is also a main component of ATP, the "cell currency" of energy transfer. All cells therefore need to maintain a sufficient concentration of phosphate in their cytoplasm and have developed systems to transport and store this nutrient. But how does a cell know how much phosphate it actually needs? Researchers from the University of Geneva (UNIGE) and the University of Lausanne (UNIL), Switzerland, report that a region of specific proteins, the so-called SPX domain, signals the phosphate status to fungal, plant and human cells. This domain provides a binding surface for small molecules that regulate the uptake of the nutrient into the cell. Their findings, which now appear in Science, could contribute to the development of crops that use phosphate more efficiently.
In order to function properly, eukaryotic cells, i.e. cells from higher living organisms, need to maintain sufficient phosphate levels. To absorb this macronutrient, fungal, plant and human cells have developed transport and storage systems. How cells know how much phosphate they contain at any given time remained however unclear. Michael Hothorn, Professor at the Department of Botany and Plant Biology of the Faculty of Science of UNIGE, and his research group revealed the crystal structure of a novel protein domain called SPX, which is involved in many phosphate signaling pathways. They discovered that SPX provides a binding surface for small compounds called inositol pyrophosphate signaling molecules (InsP), which can interact with other proteins only when they are bound to the SPX domain. As SPX domains can be attached to different proteins, such as enzymes, transporters or signaling proteins, the biologists hypothesized that InsP regulate various cell processes involved in phosphate homeostasis, from yeast to human cells.
A ubiquitous signal of the cell's phosphate status
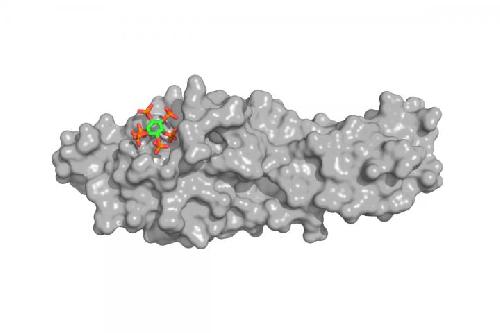 A 3-D model of the SPX domain with one InsP molecule bound. Credit: © Rebekka Wild, UNIGE
A 3-D model of the SPX domain with one InsP molecule bound. Credit: © Rebekka Wild, UNIGE
This hypothesis was explored in collaboration with researchers from the UNIL and other European universities. «We found out that the concentration of InsP changes in response to phosphate availability. InsP levels are high in cells that have sufficient phosphate, and drop when phosphate becomes scarce», explains Ruta Gerasimaite from UNIL, one of the first co-authors of the study. «In phosphate-starved plants, specific transcription factors turn on the expression of phosphate transporter genes. Once the plant is satiated, SPX domains filled with InsP will bind and inactivate these transcription factors, and no more phosphate will be absorbed from the soil into the cell», says Rebekka Wild from UNIGE, another of the first co-authors.
Yves Poirier, Professor at the Department of Plant Molecular Biology of UNIL, and his colleague Ji-Yul Jung further demonstrated this with the model plant Arabidopsis thaliana: when SPX domains present in phosphate transporters are mutated in the spot that normally binds InsP, phosphate transport is impaired.
The role of InsP was initially elucidated in yeast cells: «We came across InsP while studying the mechanism of phosphate polymerization - its assembly into long chains - for the storage of this compound, and our data show that the SPX domain is a receptor for InsP», states Andreas Mayer, Professor at the Department of Biochemistry of UNIL. Once the SPX domain is filled, it activates the enzyme involved in phosphate storage.
Better understanding of phosphate homeostasis
This research now opens up new pathways to study and better understand phosphate homeostasis in organisms and may even lead to a more efficient phosphate use in crops. «Crops on the fields are usually lacking phosphate and therefore need to be fertilized. However, the resources of phosphate fertilizer are declining worldwide. Our discovery could open the door to the development of crops that could grow efficiently on less phosphate», says Michael Hothorn.
This paper will be published online by the journal Science on THURSDAY 14 April 2016.
source: Université de Genève