Freiburg: Two metres of DNA are packed into the cell nucleus, presumably based on a strictly defined arrangement. Researchers working with biologist Patrick Heun from the Max Planck Institute of Immunobiology and Epigenetics in Freiburg have now succeeded in explaining a phenomenon, which was first observed 40 years ago. The centromeres, namely the structures at the primary constriction of the X-shaped chromosomes, are clustered in a few specific locations in the cell nucleus. Using fruit flies as a model, the researchers have shown that a single protein plays a key role in this process. If this protein is missing, DNA damage in the cell increases and cell division is impaired
Each chromosome is duplicated before cell division. However, the two copies remain connected at a single site, the centromere, until shortly before division. This gives rise to the classical X-shaped structure of chromosomes. Over the past 40 years, dye experiments have shown that in many organisms and different cell types, the centromeres of several chromosomes are clustered in the nucleus. However, it is still little understood how the DNA is organized in the nucleus.
In a series of experiments using the fruit fly Drosophila melanogaster, Patrick Heun and colleagues at the Max Planck Institute of Immunobiology and Epigenetics and associated with cluster of excellence BIOSS Centre for Biological Signalling Studies of the University of Freiburg, have now shown that the protein NLP plays a major role for centromere positioning. It binds specifically to the centromere region of the chromosomes and causes their clustering near the nucleolus, an important area of the nucleus. The site where the protein binds to the chromosome depends on the packaging of the DNA. As Heun and his team discovered, however, the DNA sequence at this site does not appear to matter. NLP also exists in a slightly modified form in humans and is known as nucleophosmin.
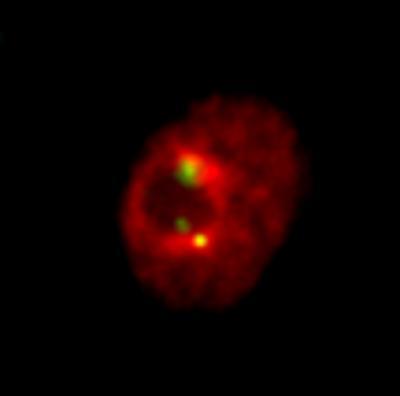
Order in the nucleus: Centromeres (marked in green) of the 13 Drosophila tissue culture chromosomes are clustered at only three positions. They attach to the nuclear region called nucleolus (dark spot). Chromatin is stained in red and marks the nucleus.
(Photo Credit: Patrick Heun, MPI of Immunobiology and Epigenetics)
If the researchers eliminate the NLP protein using the so-called gene knockdown method, the centromeres fall apart and distribute themselves throughout the nucleus. Due to this change in the spatial architecture of the nucleus, silenced areas of DNA are activated and damage accumulates in the DNA double-strand. Such changes can impair the stability of the genome and ultimately contribute to the emergence of cancer. "It is already known that the organisation of the nucleus plays a role in a specific form of leukaemia. It would be very interesting to find out whether the lack of organisation of the centromeres also plays a role in cancer cells," says Heun.
In addition to the protein NLP, the scientists also succeeded in decoding the interactions with two other already known proteins. The nucleolus protein modulo anchors the complex consisting of the centromere and NLP to the nucleolus, and the protein CTCF supports NLP in the clustering of the centromeres.
"Regarding the molecular mechanism underlying this phenomenon, we now have a foot in the door," says Heun. He has been working on the elucidation of this process with his doctoral student Jan Padeken and his team since 2008. Other studies on Drosophila and mammalian cells, for example from mice, are now expected to follow. "We anticipate that other proteins are also involved in the organisation of the centromeres and would like to find out whether the discovered protein network also plays a role in other organisms," says Heun.
Scientists at the Max Planck Institute of Immunobiology and Epigenetics (MPI-IE) in Freiburg, which was established in 1961, study how the immune system developed down through evolution and how it changes over the course of life. In 2007, an "Epigenetic Focus" was additionally established at the institute. This involves the study of the inheritance of characteristics that are not based on changes in the DNA sequence.
Source: Max-Planck-Gesellschaft